New Scientist, UK: Produced naturally by bacteria, CRISPR (clustered regularly interspaced short palindromic repeat) has gained rock-star status among scientists in the decade since its extraordinary potential was first recognised, and it is already starting to live up to the promise. But behind all the excitement lurk some dark questions. What if the editing goes wrong? What if it has undesired effects? What if we can't stop it? Without a means to keep CRISPR on target and halt it in its tracks when needed, gene editing could have disastrous consequences – both for human health and for the planet.
What we need is an off-switch, one that can be used at will. Researchers around the world have spent years trying to find one, largely by investigating various biochemical solutions. However, it turns out that the answer may be right under our noses. In an evolutionary face-off between CRISPR-producing bacteria and the viruses that infect them, nature has already designed anti-CRISPR. The challenge now is to harness this evolved off-switch to our own ends and usher in the golden age that gene editing promises.
CRISPR forms part of many bacterial genomes. It is made up of repeating DNA sequences interspersed with fragments of genetic code left behind by phages from past viral attacks. When a phage invades again, the bacterium makes RNA copies of these CRISPR regions. These bits of genetic material then hook up with a particular protein, an enzyme called Cas.
They latch on to matching sequences in the invading virus's genome, and the accompanying Cas protein snips the viral DNA strand, destroying the phage. In effect, CRISPR works as a sort of genetic memory of past viral attacks that confers immunity against future ones.
Elegant editing, Given the system's simplicity and elegance, it is perhaps unsurprising that researchers eventually spotted CRISPR's potential as a gene-editing tool. The discovery won a Nobel prize in 2020 for biochemist Jennifer Doudna at the University of California, Berkeley, and Emmanuelle Charpentier, now director of the Max Planck Unit for the Science of Pathogens in Germany.
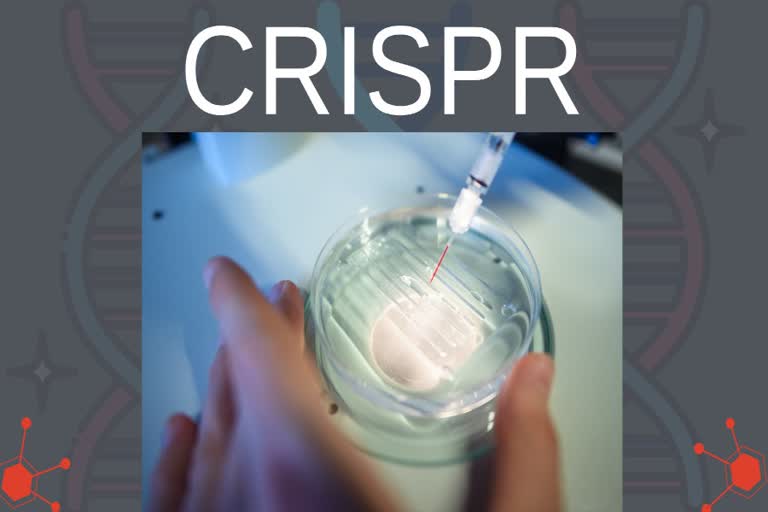
CRISPR-Cas9 has since been used successfully many times to genetically edit cells in the lab. But for it to be an effective medical therapy, it must be delivered directly to cells in the human body either physically, such as by injection, or with a vector, usually an engineered virus that encodes the desired genes.
In 2020, a team in the US achieved this for the first time, injecting CRISPR into the eyes of someone with an inherited form of blindness caused by a single mutation. Precisely targeting other parts of the body is harder, however.
The issue is how to get CRISPR only to the cells of interest, while also ensuring that enough editing takes place in them to see the changes you want.
Many scientists worry about the consequences of gene editing is left unchecked.
"Expression of Cas9 in the wrong place, or for too long, is going to be very dangerous," says microbiologist Alan Davidson at the University of Toronto, Canada.
The main problem is that CRISPR can zero in on sequences that are similar to, but not an exact match for, its target, and Cas9 is then able to cut such sequences. As a result, there is a risk that, while being used to treat a genetic disease, CRISPR-Cas9 could cause harmful changes elsewhere in a person's genome – so-called off-target edits.
Also Read: Microorganisms or microbes in our day to day life
A gene drive could be used to great effect: to eradicate a vector-borne disease such as malaria, for example, by promoting a gene that makes male mosquitoes infertile or one that prevents females from biting. But there is a danger that such genetically edited organisms might run amok in the environment with unintended consequences.

Take back control; CRISPR has been found lurking in the genomes of half of all sequenced bacteria. However, some phages have evolved their own system to fight back.
Their defence consists of small proteins called anti-CRISPRs (Acrs) encoded in their genome. When a phage infects a bacterium, it injects its genetic material and then hijacks the host's genetic machinery to make copies of its own genes.
Anti-CRISPR was discovered by accident in 2012, just as the CRISPR gene-editing revolution was taking off. In 2016, Bondy-Denomy and his colleagues found Acrs capable of disabling the Cas9 enzyme, the one used in the vast majority of gene-editing studies. His former colleague April Pawluk discovered this same protein, AcrIIA4, simultaneously.
Now CRISPR researchers did take notice. Within months, a team including Doudna had delivered AcrIIA4 into human cells along with CRISPR-Cas9, allowing them to limit gene editing to a brief period, so minimizing the problem of off-target edits. Timing when the Acr was administered provided another layer of control, allowing them to switch off Cas9 either abruptly or gradually.
For a start, it would minimize the number of therapies patients are exposed to compared with using a separate drug to inhibit Cas9. It also means you can be sure that Cas9 and AcrIIA4 are in the same place at the same time. And it can give you more control to stop and start gene editing by allowing the activity of the two proteins to be toggled back and forth – potentially by using light. Already, one group has engineered a version of AcrIIA4 that can be turned on and off by shining light onto it, a technique known as optogenetics.
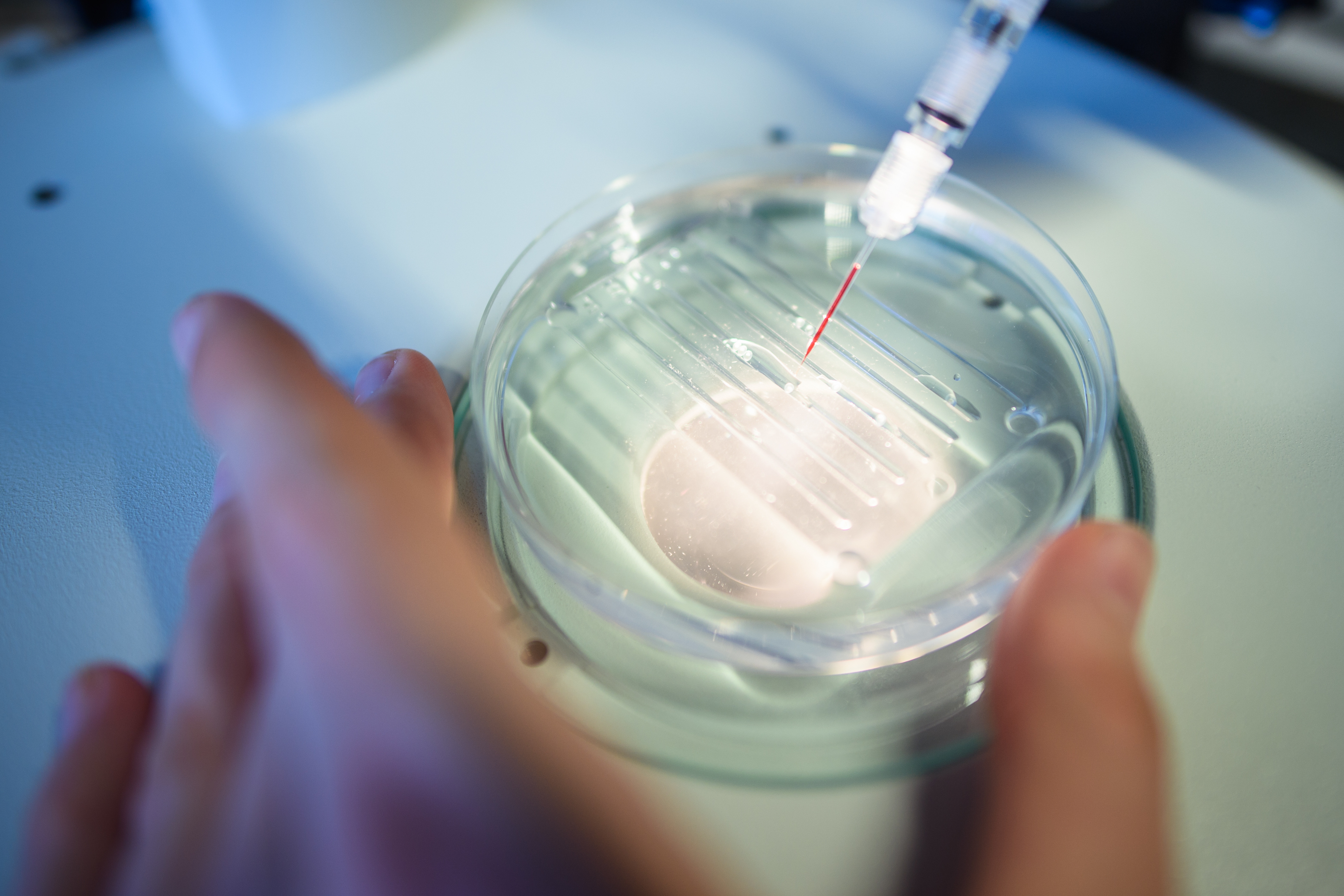
On Target; Anti-CRISPR might even help solve the problem of getting gene editing to occur only in certain cells. By tinkering with Acrs, it is possible to produce CRISPR-Cas9-Acr complexes that are permanently off in non-target parts of an organism.
Nevertheless, both Acrs and CRISPR-Cas9 are of non-human origin so could generate an immune response that would inactivate them and might cause damaging inflammation. Cas9 has already been seen to generate antibodies in mice.
However, Acrs are around 100 times smaller than Cas9, which means they stand less chance of being recognised by any antibodies that might be produced in response to them. The biggest challenge will be making anti-CRISPR work in practice.
The defence mechanism that bacteria have evolved in response to the viruses that infect them is ingenious. Known as CRISPR, it consists of two elements: a stretch of DNA that can target viruses the bacteria has encountered and an enzyme that then chops up the invaders. This ability to target and destroy has brought CRISPR to the attention of researchers aiming to develop gene editing.
However, in the natural world, CRISPR's power seems to be waning. Some bacteria-infecting viruses have evolved a protein-based counter-attack called anti-CRISPR – and it is almost always successful.
It is a bit of an evolutionary puzzle why natural selection keeps CRISPR going in bacteria that meet resistance from anti-CRISPR. One possible explanation is that CRISPR has acquired other useful functions. For example, it seems to help some bacteria form biofilms – diverse communities of microbes that have many advantages for the survival of their inhabitants. In addition, some bacteria use CRISPR to help regulate the expression of their genes. It may also have other uses yet to be discovered.
Another reason bacteria maintain CRISPR is that it is still effective against viruses with anti-CRISPR in certain circumstances. The most important factor appears to be the relative size of the virus and host populations. The CRISPR system takes energy to run, but its big advantage is that it can respond rapidly. So, when bacteria are under attack from just a few viruses, CRISPR can eliminate them before they proliferate and activate their anti-CRISPR, saving the microbes from having to expend too much energy.
(c) 2021 New Scientist Ltd.
Distributed by Tribune Content Agency, LLC
Also Read: Microbes could be used to extract metals and minerals from space rocks